StrainConfigurationHook¶
- class StrainConfigurationHook(strain_direction=None, strain_rate=None, strain_interval=1)¶
Pre-step hook apply strain to the configuration in a given cartesian direction during an MD simulation.
- Parameters:
strain_direction – The Cartesian direction to strain. May either be a uniaxial strain (e.g. “x”, “y”, “z”) or a shear strain (e.g. “xy”, “zx”, “yz”).
strain_rate (PhysicalQuantity of type inverse time) – The amount of strain per unit time applied to the chosen axis.
strain_interval (int) – The number of MD steps between when the strain is applied to the configuration.
- callInterval()¶
- Returns:
The call interval of this hook function.
- Return type:
int
Usage Examples¶
Run a 6 ps NVT MD simulation, with a strain rate of 0.25% per ps, while measuring the stress and strain at every MD step. These measurements are then used to plot a stress-strain curve.
# Define a strain configuration hook.
strain_hook = StrainConfigurationHook(
strain_direction='x',
strain_rate=0.005 / ps,
strain_interval=1000,
)
measurement_names = ['stress_xx', 'strain_xx']
md_trajectory = MolecularDynamics(
bulk_configuration,
constraints=constraints,
trajectory_filename='stress_strain.hdf5',
steps=6000,
log_interval=1000,
pre_step_hook=strain_hook,
measurement_hook=MDMeasurement(measurement_names, call_interval=1),
method=method,
)
bulk_configuration = md_trajectory.lastImage()
# Get stress strain values.
times, strains = md_trajectory.measurement('strain_xx')
times, stresses = md_trajectory.measurement('stress_xx')
# Perform linear fit.
polynomial_coefficients = numpy.polyfit(strains, stresses.inUnitsOf(GPa), 1)
youngs_modulus = polynomial_coefficients[0] * GPa
# Evaluate polynomial to plot best fit line.
stresses_fit = numpy.polyval(polynomial_coefficients, strains)
# Plot the fit.
import pylab
pylab.plot(
strains,
stresses_fit,
label="Linear Fit (Young's Modulus=%.1f GPa)" % youngs_modulus,
)
# Plot MD data.
pylab.scatter(
strains,
stresses.inUnitsOf(GPa),
label='MD Data',
color='k',
)
# Setup plot.
pylab.xlabel('Strain')
pylab.ylabel('Stress (GPa)')
pylab.legend()
# Save to file.
pylab.savefig('stress_strain.png')
The resulting plot will look like:
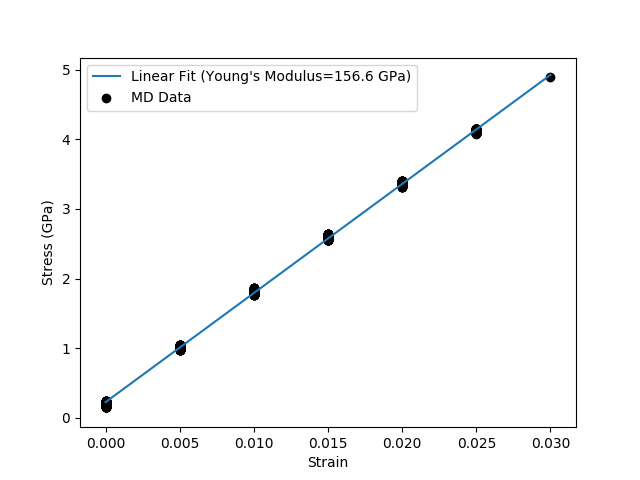
The full script can be downloaded: stress_strain.py
Stress Strain Calculations¶
In a stress-strain calculation the cell is extended or strained in one
direction and the resulting stress in that direction is then recorded. In the
elastic region, the stress ideally varies linearly with the strain. The
derivative of the stress-strain curve is the Young’s modulus of the material.
The StrainConfigurationHook enables you to easily calculate the
Young’s modulus by extending the cell during the simulation in the specified
direction. The stress during the molecular dynamics run can then be recorded
using an MDMeasurement object. The StrainConfigurationHook is
included in the molecular dynamics simulation by using the pre_step_hook
argument. This argument takes functions or callable objects that are called
before the coordinates are integrated in each molecular dynamics step.
Notes¶
The strain is defined as the engineering strain, which is the relative change from the initial unit cell.
For additional information on using hooks in molecular dynamics see the notes section of the MolecularDynamics reference manual entry.