EndToEndDistances¶
- class EndToEndDistances(md_trajectory, polymer_tags=None, end_tags=None, start_time=None, end_time=None, time_resolution=None, gui_worker=None)¶
Create an analyzer to calculate the end to end distance of polymer chains.
- Parameters:
md_trajectory (
MDTrajectory|AtomicConfiguration) – The MDTrajectory or AtomicConfiguration that contains the polymers.polymer_tags (list of type str | None | Automatic) – A list of tags for the polymers. If not specified default tags are used. Tags can also be set automatically with the flag Automatic
end_tags (list of type str | None) – A list of tags for the ends of each polymer chain. If not specified default tags are used.
start_time (PhysicalQuantity of the type time) – The start time. Default:
0.0 * fs.end_time (PhysicalQuantity of the type time) – The end time. Default: The last time image.
time_resolution (PhysicalQuantity of type time) – The time interval between snapshots in the MD trajectory that are included in the analysis.
gui_worker (PolymerAnalysisWorker) – Handle to the worker, for displaying the calculation progress. Default: None.
- autocorrelation()¶
- Returns:
The end to end vector autocorrelation function. This is returned as a 2D array indexed by polymer chain and then lag time.
- Return type:
PhysicalQuantity of type length
- distances()¶
- Returns:
The end to end distance for each polymer chain. This is returned as a 2D array indexed by trajectory image then polymer chain.
- Return type:
PhysicalQuantity of type length
- polymerTags()¶
Return the polymer tags used for the analysis.
- Returns:
A list of the polymer tags.
- Return type:
list
- times()¶
- Returns:
Times for each image used in the analysis.
- Return type:
PhysicalQuantityof type time
- vectors()¶
- Returns:
The end to end vector for each polymer chain. This is returned as a 3D array indexed by trajectory image, polymer chain then vector components.
- Return type:
PhysicalQuantity of type length
Usage Examples¶
Calculate the end-to-end distance and distance vector autocorrelation function of polystyrene chains during a molecular dynamics simulation starting from a well equilibrated configuration.
# -------------------------------------------------------------
# Initial Configuration
# -------------------------------------------------------------
bulk_configuration = nlread('Polystyrene.hdf5')[-1]
# -------------------------------------------------------------
# OPLS-AA Calculator
# -------------------------------------------------------------
potential_builder = OPLSPotentialBuilder()
calculator = potential_builder.createCalculator(bulk_configuration)
bulk_configuration.setCalculator(calculator)
# -------------------------------------------------------------
# Molecular Dynamics
# -------------------------------------------------------------
initial_velocity = ConfigurationVelocities(
remove_center_of_mass_momentum=True
)
method = NPTMartynaTobiasKlein(
time_step=1*femtoSecond,
reservoir_temperature=300*Kelvin,
reservoir_pressure=1*bar,
thermostat_timescale=100*femtoSecond,
barostat_timescale=500*femtoSecond,
initial_velocity=initial_velocity,
heating_rate=0*Kelvin/picoSecond,
)
constraints = [FixCenterOfMass()]
md_trajectory = MolecularDynamics(
bulk_configuration,
constraints=constraints,
trajectory_filename='Polystyrene_Trajectory.hdf5',
steps=100000,
log_interval=10,
trajectory_interval=1000,
method=method
)
# -------------------------------------------------------------
# Calculate End to End Distance
# -------------------------------------------------------------
analyzer = EndToEndDistances(
md_trajectory,
start_time=0*ps,
end_time=100*ps,
end_tags=['HEAD_CONNECT', 'TAIL_CONNECT']
)
end_end_distance = analyzer.distances()
end_end_distance_average = end_end_distance.mean(axis=1)
end_end_correlation = analyzer.autocorrelation().mean(axis=0)
time = analyzer.times().convertTo(ps)
# -------------------------------------------------------------
# Plot The Results
# -------------------------------------------------------------
import pylab
pylab.figure()
pylab.plot(time, end_end_distance[:, 0], label='Polymer 1')
pylab.plot(time, end_end_distance[:, 5], label='Polymer 6')
pylab.plot(time, end_end_distance_average, label='Average')
pylab.xlabel('Time / ps')
pylab.ylabel('End to end distance / Angstrom')
pylab.legend()
pylab.savefig('End_To_End_Distances_Plot.png')
pylab.figure()
pylab.plot(time[:51], end_end_correlation[:51])
pylab.xlabel('Time / ps')
pylab.ylabel('End to end autocorrelation')
pylab.savefig('End_To_End_Autocorrelation_Plot.png')
EndToEndDistances_Example.py
Polystyrene.hdf5
The script will run a short molecular dynamics calculation from the given starting structure and then calculate the end to end distances of two individual polymers, as well as the average. The end to end vector autocorrelation is also calculated. These are shown in the following two plots.

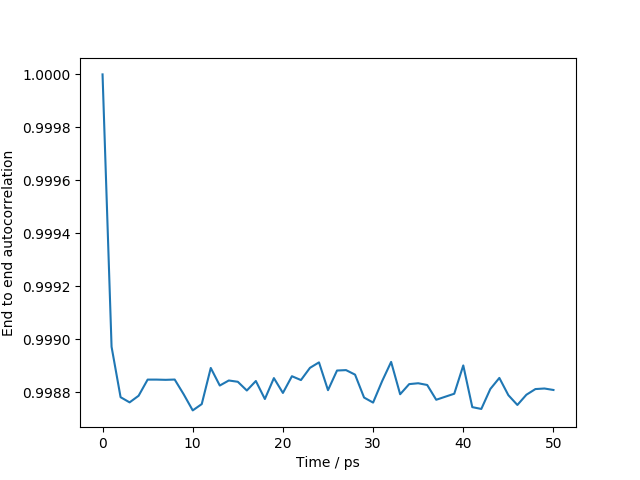
Note that the high value of the end to end vector autocorrelation indicates that there is little overall movement in the polymer chains, as the configuration is below the glass transition temperature.
Notes¶
This class enables the calculation of the end-to-end distances of different polymer chains, as well
as their autocorrelation functions from a MDTrajectory or BulkConfiguration.
The end-to-end distance is simply defined as the Cartesian distance between the two end backbone atoms in the polymer chain. For this calculation, the polymer chain is unwrapped from the simulation cell and considered as one complete molecule. Given that this distance is a vector quantity, an autocorrelation function can be calculated that measures the evolution of the direction of this vector. The end-to-end autocorrelation function \(R_{ee}(t)\) can be defined as:
To calculate each of these shape measures a EndToEndDistances object first needs to be
created. This takes a MDTrajectory or BulkConfiguration and calculates the end
to end distances and autocorrelation function for the specified configurations. The
specific frames in the MDTrajectory to analyzed can be selected using the
start_time, end_time and time_resolution arguments. The polymer chains analyzed in the
configuration can be specified with the polymer_tags argument. This takes a list of tags of
polymer molecules in the configuration. By default polymers tagged with POLYMER_MOLECULE_# are
analyzed, where # is a numerical index. Tags indicating the start and end of each polymer
chain also need to be added. The argument end_tags takes a list of strings that contain the
tags for the end atoms. These tags can be specified in any order. By default the tags
HEAD_CONNECT and TAIL_CONNECT are used. All the default polymer and end tags are placed on
polymer configurations built with the PolymerMonteCarloBuilder.
Once the EndToEndDistances object is created and the values of each of the descriptors
has been calculated, the values can be queried using the remaining methods. The method
vectors() returns a list of the end-to-end vectors, indexed by trajectory
image, then polymer chain and finally vector components. Similarly the method
distances() returns a list of the scalar end-to-end distances, indexed by
trajectory image and then polymer chain. The method autocorrelation()
returns a list of the autocorrelation functions, indexed first by polymer chain and then by lag
time.
